GPU-Accelerated Drug Discovery with Docking on the Summit Supercomputer: Porting, Optimization, and Application to COVID-19 Rese
GitHub - purnawanpp/Docking-4ieh: Docking Tutorial Using Autodock Vina version 1.2.3 (2021) and AutoDock-GPU Version 1.5.3
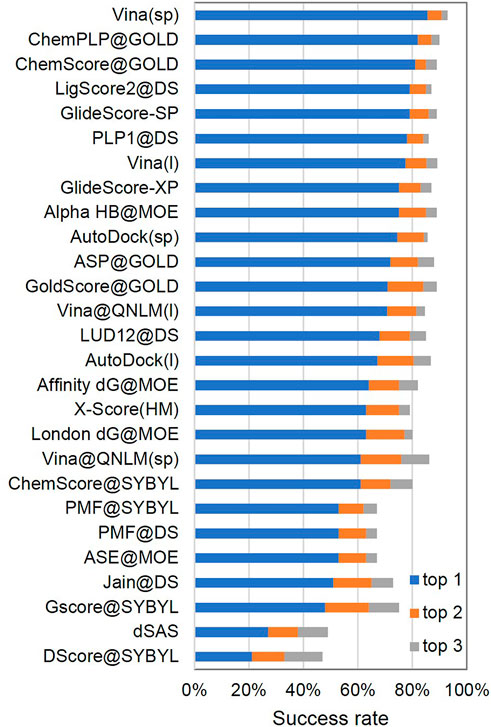
Frontiers | Redesigning Vina@QNLM for Ultra-Large-Scale Molecular Docking and Screening on a Sunway Supercomputer

Benchmarking the performance of irregular computations in AutoDock-GPU molecular docking - ScienceDirect

Docking scores of VINA-GPU on six PDB structures with various levels of... | Download Scientific Diagram
![PDF] AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings | Semantic Scholar PDF] AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e85c80d197a26c92b844ac25aedbfd71a59d16e1/2-Table1-1.png)
PDF] AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings | Semantic Scholar